Newsroom
#LabLifeLessons: A Brief History of Discovery – How Applied Science Relies on Basic Research
Written by Zack Gerbec from the Finlay lab, Michael Smith Laboratories
“A LOT OF PRIZES HAVE BEEN AWARDED FOR SHOWING THE UNIVERSE IS NOT AS SIMPLE AS WE MIGHT HAVE THOUGHT!”
-STEPHEN HAWKING, A BRIEF HISTORY OF TIME
While every research project is built with the goal of one day making the transition from basic research to applied science, most scientific discoveries will not make an immediate impact beyond the lab. Though this may seem like a counterproductive or at the very least inefficient system, those discoveries that do make the leap into the real world are invariably built on a foundation of basic science discoveries that at the time, made little impression outside of their field. As part of our #LabLifeLessons series, we want to recognize these discoveries by telling the stories of some of the basic research studies that formed the blueprint for the applied science work being done here at Michael Smith Laboratories. In some cases, our investigators were integral parts of the initial discoveries that buoyed their later work. In all cases, the real world benefits of the research being done at the Michael Smith Laboratories today would not be possible without the basic science discoveries that at one point may have only served to make things less simple than we might have thought.
Story 1: From subatomic particles to international agriculture
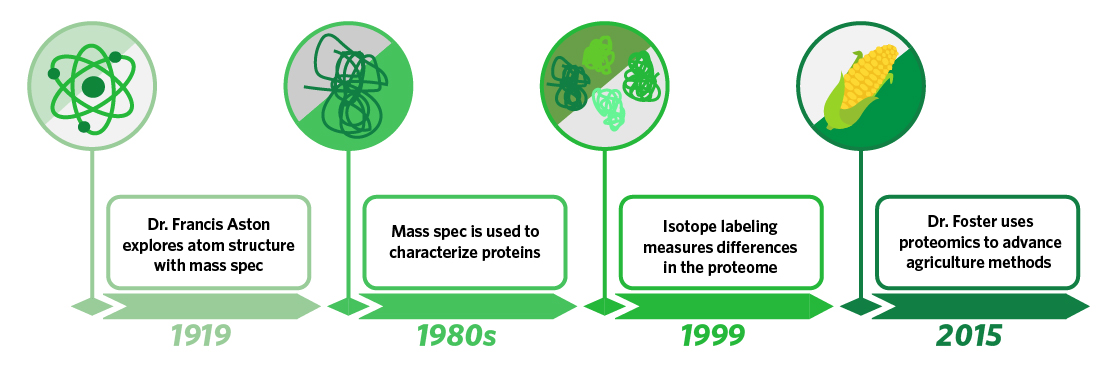
While researchers today are trying to learn how best to use microscopic molecules as tools, scientists were first trying to learn what these molecules were made of. In 1919, Dr. Francis Aston built a mass spectrometer capable of separating atoms of the same element that had uniform charges but different masses, providing a powerful tool in evaluating the subatomic particles that made up atoms. Though this was not the invention of mass spectrometry, it is one of the best early examples of the technology being used to delineate atomic structure, and it paved the way for biologists in the 1980s to develop the technology to use mass spectrometry to determine the mass of proteins. In 1999, Dr. Brian Chait developed a method of isotopically labeling proteins from different samples. This advancement enabled comparison of protein abundance in different samples, and the modernization of proteomics meant a technology that was originally used to determine atomic structure could now be used to evaluate differences in biological function. In 2015, Dr. Leonard Foster of the Michael Smith Laboratories applied isotopic labeling and proteomics technology to research on the Western honey bee and used mass spectrometry to identify bees harboring a specific proteome that inferred increased resistance to a deadly parasite. Using this information, Dr. Foster was able to use selective breeding to create a resistant strain of bees with improved survival traits. Given the critical role that honey bees play in pollinizing a wide variety of crops (the estimated contribution to agriculture is over $10 billion between Canada and the U.S.) this research has significant implications for how the world’s food supply will be maintained in the future.
Story 2: Natural tools are the best tools
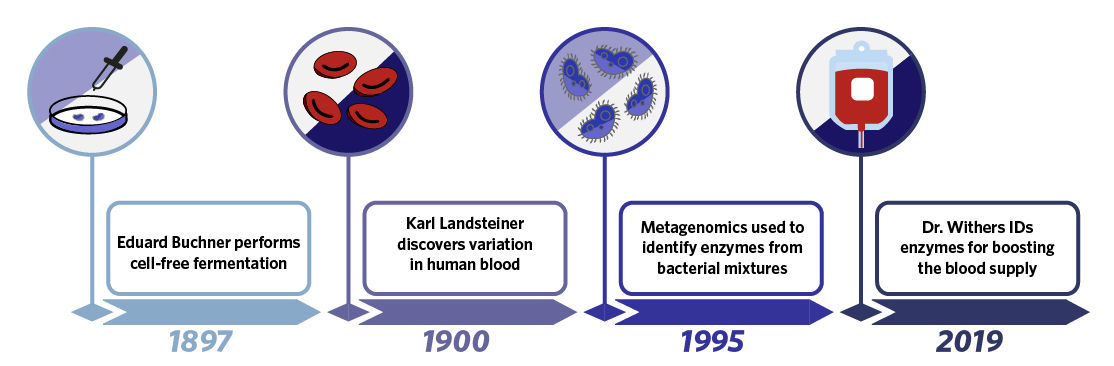
In 1897, German chemist Eduard Buchner discovered cell-free fermentation when he found that extracts from yeast were capable of fermenting sugar, demonstrating the utility of enzymes on their own and the concept of isolating enzymes with specific functions from living organisms. Three years later in a seemingly unrelated discovery, Karl Landsteiner found that an individual’s blood would agglutinate (clump) when mixed with the blood of other individuals, leading to the modern system of blood typing. At the time of this discovery, it was not known that the different blood types resulted from carbohydrate antigens present on red blood cells, and that the presence of these carbohydrates meant blood types could be modified using the cell-free enzymology concepts Buchner had demonstrated. In 1995, in a third seemingly unrelated discovery, Dr. KT Shanmugam created the first metagenomic library where DNA from a mixture of organisms was fragmented and expressed in E. coli to enable identification of single genes or small subsets of genes with specific functions. This original library was used to identify enzymes capable of breaking down cellulose, and set a precedent for utilizing metagenomics as a tool for unbiased enzyme discovery. In 2019, Michael Smith Laboratories professor Dr. Stephen Withers would use a similar screening concept to identify enzymes from human bacterial samples capable of modifying blood group carbohydrates. From his screen and in a cell-free enzymology setting, Dr. Withers was able to identify a pair of enzymes capable of efficiently converting Type A red blood cells to Type O red blood cells. By combining concepts demonstrated by three basic science discoveries, Dr. Withers was thus able to identify a new tool for increasing the world’s pool of universal blood donors.
Story 3: From an unknown protein to better brain treatments
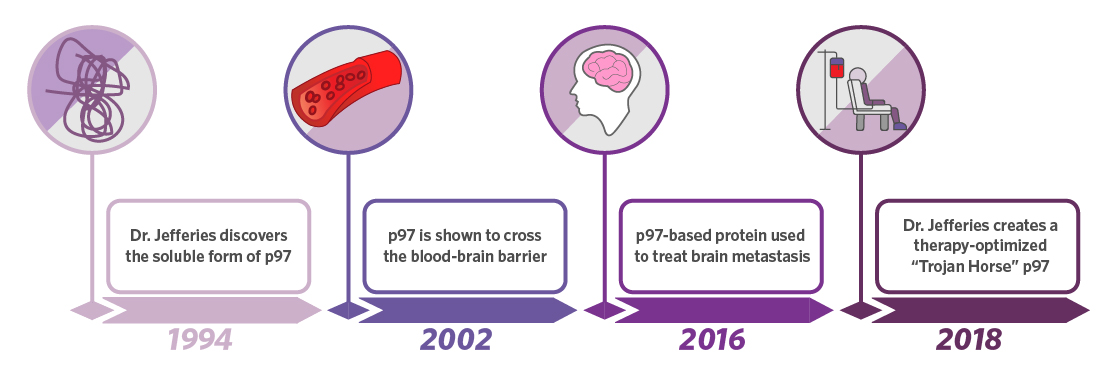
In the 1980s and 90s, Dr. Wilfred Jefferies of the Michael Smith Laboratories played a critical role in defining the structure and function of transferrin receptors on endothelial cells that move iron across the blood brain barrier. In 1994, he discovered that a soluble form of melanotransferrin existed that was not anchored to the plasma membrane. The function of this soluble protein was unknown until 2002 when it was demonstrated that the protein could actually cross the blood brain barrier itself rather than just transport iron. The utility of this was not lost on the biomedical field, and the therapeutic potential of a brain delivery system was tested in multiple disease models including glioblastoma in 2016. The success of these pre-clinical models enabled Dr. Jefferies to invent an optimized melanotransferrin peptide capable of delivering drugs across the blood brain barrier. This optimized delivery system, which would not exist without the initial discovery made 26 years ago, has been used successfully for treatment in multiple pre-clinical models including those for neuropathic pain and stroke.
Story 4: A little more than arts and crafts
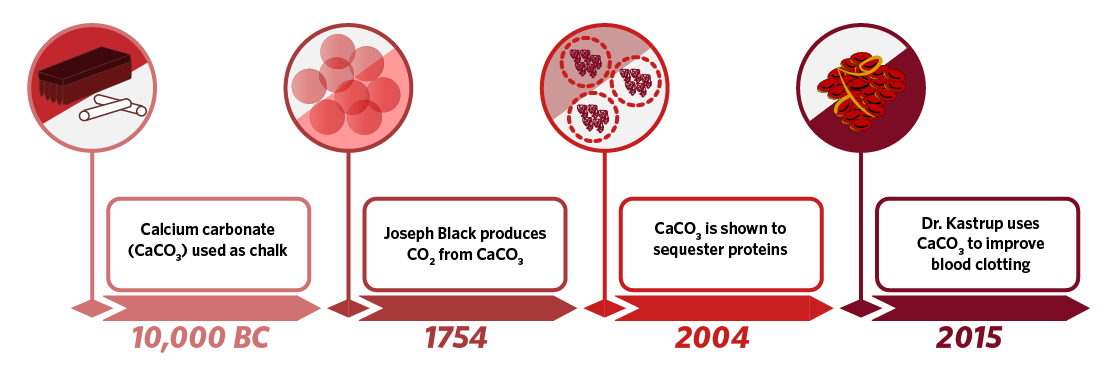
The first use of calcium carbonate was not in a test tube or a culture dish, but rather as chalk to make cave drawings some time before 10,000 BC. Much later in 1754, the Scottish chemist Joseph Black found that when calcium carbonate was heated or treated with acid, it produced a gas that he called fixed air. The gas turned out to be carbon dioxide, and was the first example of a now well-established chemical reaction in which decomposition of a metal carbonate produces a metal oxide and carbon dioxide. In 2004, further characterization of calcium carbonate revealed that synthesized microparticles could encapsulate and stably sequester proteins. Taking advantage of this discovery, Michael Smith Laboratories investigator Dr. Christian Kastrup found that these microparticles could take up and hold significant quantities of thrombin, the protein that drives coagulation and blood clotting. He also correctly hypothesized that by adding a solid organic acid to the mixture, he could recapitulate the chemical reaction first demonstrated by Joseph Black and that the particles would produce carbon dioxide. In synthesizing thrombin-loaded particles capable of producing gas in solution, Dr. Kastrup created a delivery system that in theory and in practice could travel up through a blood vessel and initiate clotting. Dr. Kastrup was able to demonstrate the effectiveness of these particles in multiple animal models of severe hemorrhage, and, through correct implementation of the basic properties of calcium carbonate, create a molecule that could potentially be used both in a trauma setting and as a way to enhance drug delivery.
Why do you think basic research is important? Join the conversation on Twitter with @ubcmsl with #lablifelessons.
_______
This article is part of the #LabLifeLessons blog series, a series that highlights the adventures of the PhD experience and beyond. Written by Postdoctoral Fellows at the Michael Smith Laboratories, this series includes a number of posts ranging from personal experiences, interviews, and stories, reflecting on the journey and extracting the learned lessons in the process. #LabLifeLessons focuses on these challenges and aims to bring an authentic voice to the story. If you enjoyed this story, check out the other articles below.
_______