Newsroom
Dr. Michael Skinnider receives Bayer Foundation Early Excellence in Science Award
 We extend our enthusiastic congratulations to Dr. Michael Skinnider for receiving the Bayer Foundation Early Excellence in Science Award in Data Science. This competition aims to “…identify the rising stars of science across the globe in four disciplines: biology, chemistry, medical science, and data science in life sciences”.
We extend our enthusiastic congratulations to Dr. Michael Skinnider for receiving the Bayer Foundation Early Excellence in Science Award in Data Science. This competition aims to “…identify the rising stars of science across the globe in four disciplines: biology, chemistry, medical science, and data science in life sciences”.
An exceptional computational biologist, holding both an MD and PhD from the University of British Columbia, Dr. Skinnider recent joined the Ludwig Princeton Branch and the Lewis-Sigler Institute for Integrative Genomics at Princeton University as an assistant professor. Formerly a member of the Foster lab at the MSL, Dr. Skinnider (he/him) focused his laboratory doctoral work on developing computational tools for mass spectrometry-based proteomics. Upon completing his combination MD and PhD studies, he decided to focus solely on the laboratory aspect of his work.
“Creativity has such a profound impact in science, perhaps more so than in medicine. One of the goals of clinical practice guidelines, for example, is to try and standardize the delivery of healthcare. And in science it’s the complete opposite. You’re really only limited by your ability to imagine new technologies and try to put them into practice. This is really attractive to me. I feel by focusing on laboratory science I can bring a more unique skillset to the table and make a bigger impact,” reflects Skinnider.
That innovative skillset and creative problem solving has landed Skinnider his position at Princeton University where his research will expand upon some groundbreaking tools he created to better understand small molecules, which are foundational to many larger building blocks of science and life.
“We can do amazing things with proteomics, genomics and transcriptomics to measure DNA, RNA and proteins in a given sample. But there is a tremendous gap with our ability to measure small molecules, it’s essentially impossible. This is an enormous part of our world that we have no ability to understand. Small molecules make up such a large part of our world and experience. It’s staggering to think about how little we know about them,” Skinnider explains.
Having previously worked on the discovery and analysis of small molecules gathered from the environment that could be used as candidates for antibiotics, Skinnider was inspired by how immediately translatable this science was for medical applications. At the nascence of deep learning technology development, during Skinnider’s PhD studies in the Foster lab the team created an artificial intelligence tool called DeepMet. This is used for small molecule identification and analysis in great detail. Known as metabolomics, the information gathered by this method can identify molecules associated with disease, produce diagnostic and prognostic tests and help inform patient/drug interactions.
“I’m really excited about what the next steps will be with the technology and our ability to shine a light on this previously unknown chemistry within our body,” says Skinnider.
Skinnider’s lab is building new computational tools that create a textual representation of small molecules’ chemistry, representing their make up as language. They then train deep neural networks similar to ChatGPT, originally developed to parse human language, on this textual representation of the chemistry. These models can both generate new molecules and predict which molecules will be discovered in the future. This technology has forensic chemistry applications and can be used to identify emerging ‘designer drugs’, synthetic opioids, cannabinoids and stimulants, from patient samples. The BC Centre for Disease Control is now piloting a drug identification program based on these techniques to assist with the drug poisoning epidemic in British Columbia, with the goal of improving care and speeding up the discovery of new designer drugs. This technology can also be used predictively to anticipate novel drug recipes.
“Trained on previously discovered designer drugs, these computational models can predict the drugs that clandestine chemists are mostly likely to make next,” explains Skinnider.
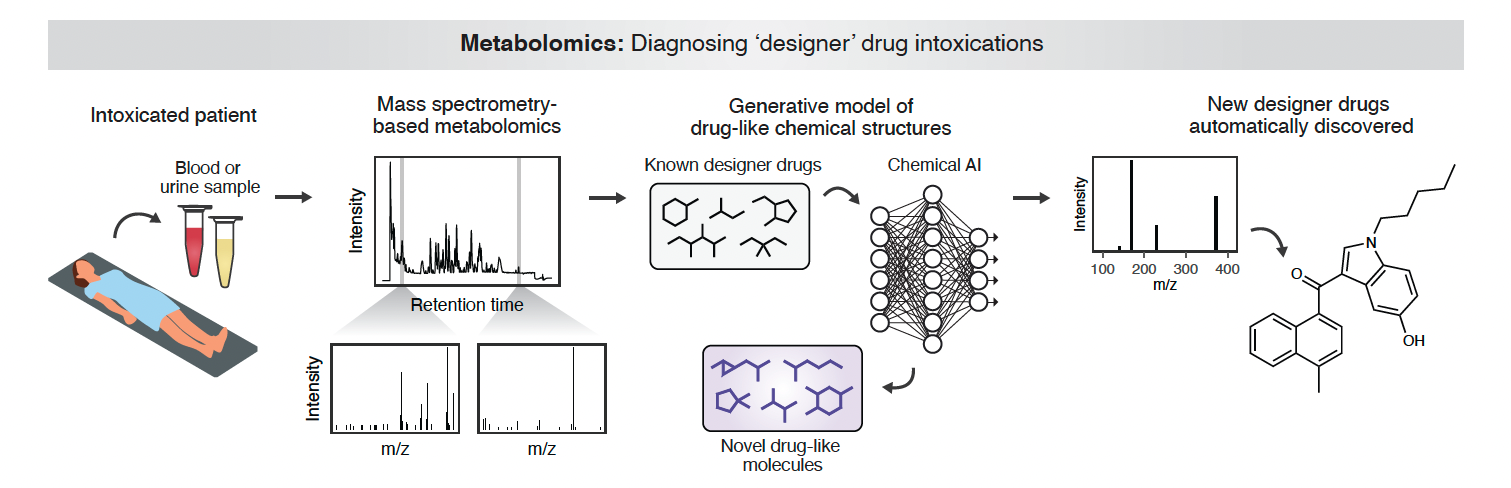
Dr. Skinnider’s ultimate vision for the future of this research is to be able to take any biological sample, analyze it with mass spectrometry, and with the data collected capture a complete list of all the known and unknown small molecules in that sample. This would allow for a holistic look at these samples and their make-up, rather than analyzing things in isolation. Current methods can only analyze about two to ten percent of the small molecules in a given sample while linking their proteomics to their chemical structure.
“Our ability to solve this problem has hit a plateau. We need to figure out how to pull more out of this data in order to get better at this problem of solving chemical structures. And the way to do that is to delve further into sources of data that are being overlooked. What’s exciting to me as a computation biologist is that this gap is purely computational,” emphasizes Skinnider.
Another exciting direction of this research is exploring the connection between the human gut microbiome and cancer. Small molecules in the gut can promote the growth of cancer, protect against it or affect the treatment of it. The myriad of therapeutic applications for this technology are inspiring.
Humbled by his nomination, Dr. Skinnider is grateful to have his groundbreaking work acknowledged in this way.
“The computational biologists that have been nominated for this award previously are people I really look up to and admire their work. To be nominated alongside them is truly an honour,” he says.
We are optimistic about how Dr. Skinnider’s novel research will propel the scientific community past the field’s current hurdles. And we applaud him for this noteworthy achievement.
Quick links:
- Bayer Foundation announcement
- Learn more about Dr. Leonard Foster’s lab
- More about Dr. Michael Skinnider’s research