Newsroom
Bioinformatics tool tracks genomic variants found in individuals with Autism Spectrum Disorder
According to a recent report by the Public Health Agency of Canada, each person with Autism Spectrum Disorder (ASD) is unique and will have different symptoms, deficits, and abilities. This range of characteristics spans a “spectrum”, in which each individual’s abilities and deficits vary widely.

Manuel Belmadani, Bioinformatician and first author of the paper.
Research on ASD has benefited from new technology such as whole genome sequencing, leading to an influx of data on genomic variants found in individuals with ASD. Although these vast amounts of collected data have benefited research, the inconsistencies in methodology and subject overlap across studies complicate the data, making it difficult for researchers to access and use. VariCarta, a new online database created by researchers in the laboratory of Dr. Paul Pavlidis, collects ASD data in a carefully curated and consistent way that is easy-to-use and is accessible to all researchers.
The database is described in a recently published paper in Autism Research Journal.
“Collecting data from diverse sources comes with a lot of challenges such as properly resolving duplicates, catching reporting errors, and simply making sense of cases where two sources are saying different things,” says Manuel Belmadani, Bioinformatician and first author of the paper. “Having done that work for our own ASD genetics research projects, we thought VariCarta would be a helpful resource for other researchers.”
VariCarta currently represents the largest collection of systematically curated, harmonized, and annotated literature-derived variants specific to ASD. With over 170,000 genomic variants found across approximately 11,000 ASD individuals, VariCarta offers a comprehensive resource of precisely characterized variants that can be queried in different ways to produce a subset of variants of interest.

Dr. Sanja Rogic, Research Associate in the laboratory of Dr. Paul Pavlidis and corresponding author of the paper
“We provide visualizations, search tools, and various annotations, as well as, group matching reports from different sources,” explains Belmadani. “The downloaded data set provides an easy way for researchers to take the curation and harmonization work and apply it to their own research questions.”
Not only is VariCarta user-friendly, it provides a more consistent way to analyze ASD genomic data, a result that came out of the researchers’ own experience when studying ASD. During their research, they found that it was difficult to asses whether a gene had already been reported in other studies or if cohorts used in different projects came up with the same results. They found that the analysis and reporting of variants across studies can vary, making it difficult to compare across papers.
“Double-reporting of variants in the literature can lead to inflation of gene-based evidence for ASD association, which often relies on the observation of likely damaging variants, particularly de novo ones, recurring in the same gene in unrelated individuals,” explains Dr. Sanja Rogic, Research Associate in the laboratory of Dr. Paul Pavlidis and corresponding author of the paper. Rogic emphasizes that variant harmonization was an important aspect of the database’s curation process, identifying and linking together unique variants that have been reported in literature multiple times due to patient cohort overlaps, re-sequencing and re-analysis. When asked how VariCarta differs from other ASD databases, Rogic states that VariCarta uses variant harmonization “better than other ASD databases” and “aggregates data across more publications than most other databases”. Members of Dr. Paul Pavlidis’ laboratory aims to continue work in ASD-related research projects and hopes databases such as VariCarta will make genetics research more accessible across the research community.
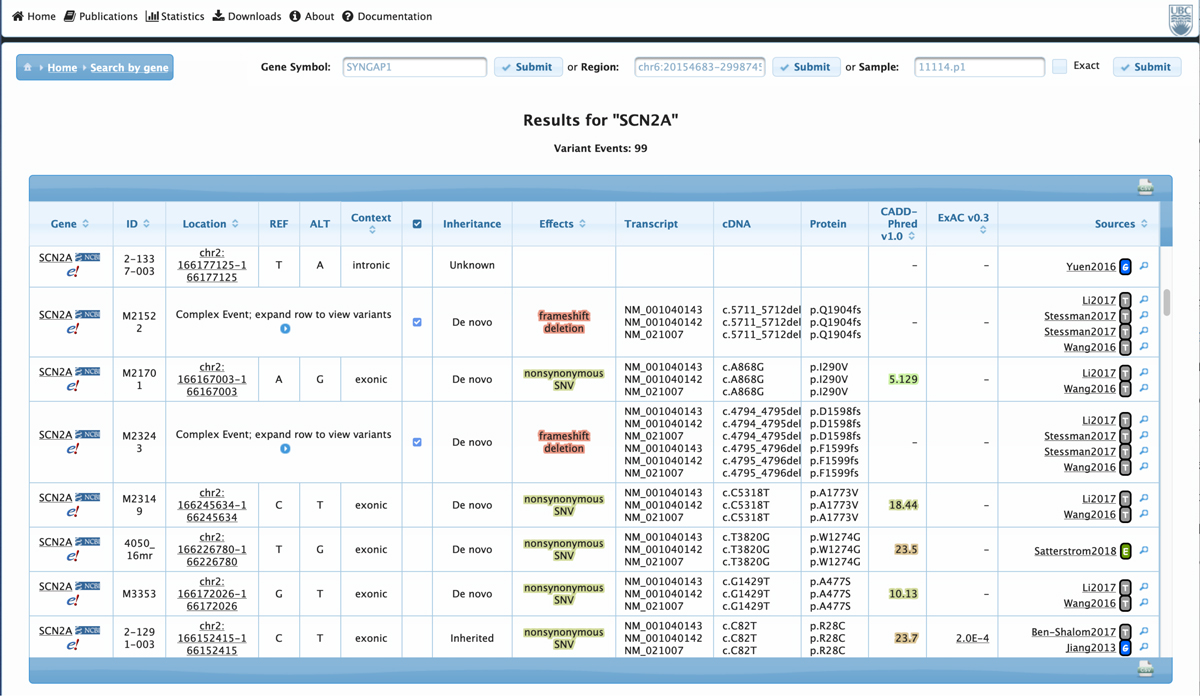
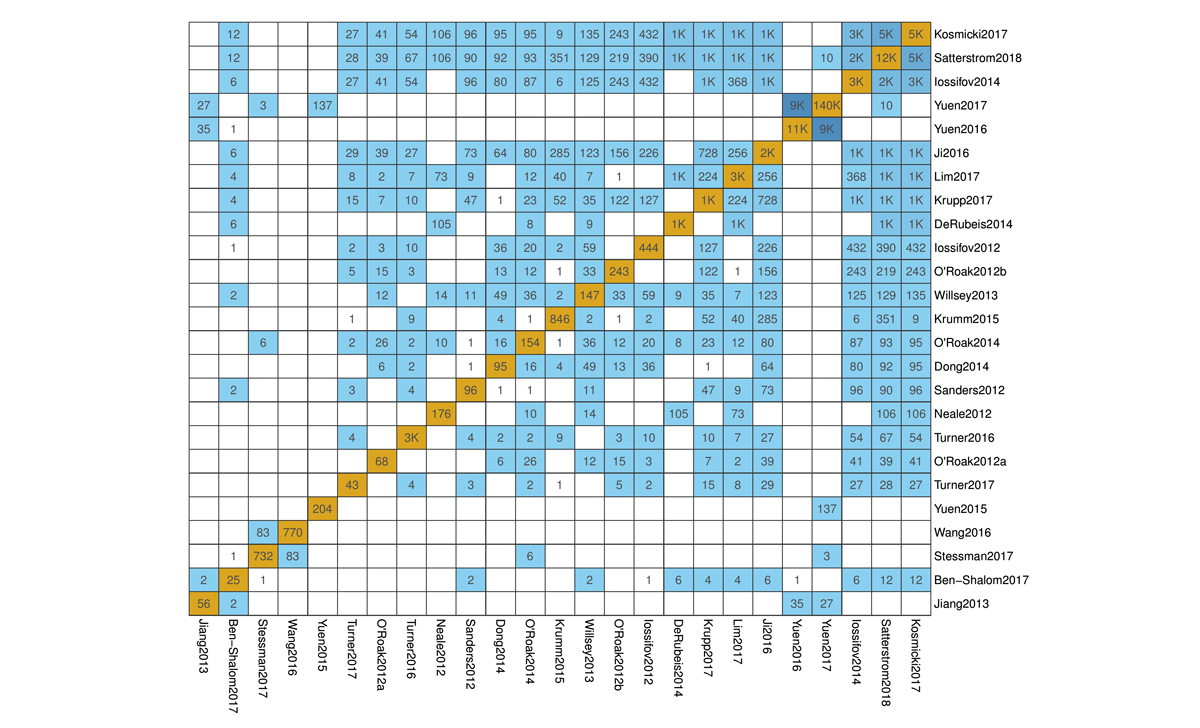
Pictured are screenshots of the VariCarta database. On the left, a resulting variant table for the SCN2A gene and on the right, an image showing the extent of variant overlap between pairs of papers in the database (click to enlarge). The VariCarta database is free to use for academic and non-commercial purposes and can be found at varicarta.msl.ubc.ca.
This research was funded by the Simons Foundation.